What’s wrong with root mean squared error for logistic regression?#
Show code cell content
import numpy as np
import pandas as pd
# Safe setting for Pandas. Needs Pandas version >= 1.5.
pd.set_option('mode.copy_on_write', True)
import matplotlib.pyplot as plt
# Make the plots look more fancy.
plt.style.use('fivethirtyeight')
# Optimization function
from scipy.optimize import minimize
This page gives some extra explanation for the logistic_regression page.
Here we say more about why we might prefer the Maximum Likelihood Estimate way
of scoring potential fits to the data, to our more usual least squared error.
If you want the gory details on this choice, see this answer on
StackOverflow. Here we look at
whether this the root mean squared error works well with minimize. The
discussion in this page corresponds to the “computational efficiency” section
of the answer linked above.
The background, the data#
In that page we were trying to looking at the chronic kidney disease dataset, to see if we good predict whether a patient had “good” appetite (as opposed to “poor” appetite) given that patient’s blood hemoglobin concentration.
Show code cell content
df = pd.read_csv('ckd_clean.csv')
# Our columns of interest.
hgb_app = df.loc[:, ['Hemoglobin', 'Appetite']]
# Dummy value column containing 0 for "poor" Appetite, 1 for "good".
hgb_app['appetite_dummy'] = hgb_app['Appetite'].replace(
['poor', 'good'],
[0, 1])
hgb_app.head()
/tmp/ipykernel_5538/1768524625.py:5: FutureWarning: Downcasting behavior in `replace` is deprecated and will be removed in a future version. To retain the old behavior, explicitly call `result.infer_objects(copy=False)`. To opt-in to the future behavior, set `pd.set_option('future.no_silent_downcasting', True)`
hgb_app['appetite_dummy'] = hgb_app['Appetite'].replace(
| Hemoglobin | Appetite | appetite_dummy | |
|---|---|---|---|
| 0 | 11.2 | poor | 0 |
| 1 | 9.5 | poor | 0 |
| 2 | 10.8 | poor | 0 |
| 3 | 5.6 | poor | 0 |
| 4 | 7.7 | poor | 0 |
We take out the columns we are interested in for our further use:
# The x (predictor) and y (to-be-predicted) variables.
hemoglobin = hgb_app['Hemoglobin']
appetite_d = hgb_app['appetite_dummy']
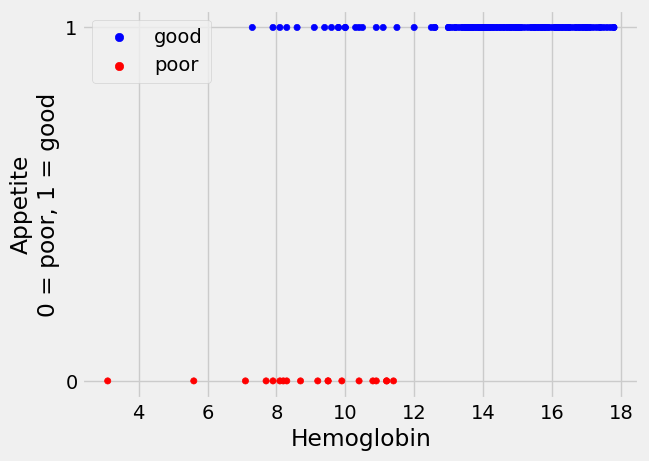
Here is a plot of the 0 / 1 appetite values
Show code cell content
def plot_hgb_app():
# Build plot, add custom label.
colors = hgb_app['Appetite'].replace(['poor', 'good'], ['red', 'blue'])
hgb_app.plot.scatter('Hemoglobin', 'appetite_dummy', c=colors)
plt.ylabel('Appetite\n0 = poor, 1 = good')
plt.yticks([0,1]); # Just label 0 and 1 on the y axis.
# Put a custom legend on the plot. This code is a little obscure.
plt.scatter([], [], c='blue', label='good')
plt.scatter([], [], c='red', label='poor')
plot_hgb_app()
plt.legend();
Linear regression - the crude approach#
The crude and brutal approach to predicting these values is to use simple
least-squares regression. We can do this in the usual way by using
scipy.optimize.minimize with a function that returns the root mean squared
error between the straight line predictions and the 0 / 1 labels. Here’s the
function from the Using minimize page.
def rmse_any_line(c_s, x_values, y_values):
# c_s is a list containing two elements, an intercept and a slope.
intercept, slope = c_s
# Values predicted from these x_values, using this intercept and slope.
predicted = intercept + x_values * slope
# Difference of prediction from the actual y values.
error = y_values - predicted
# Root mean squared error.
return np.sqrt(np.mean(error ** 2))
Start with a guess of intercept -0.5, slope 0.1:
# Use cost function with minimize.
mr_ss = minimize(rmse_any_line, [-0.5, 0.1], args=(hemoglobin, appetite_d))
mr_ss
message: Optimization terminated successfully.
success: True
status: 0
fun: 0.2555152342856677
x: [-7.904e-02 7.005e-02]
nit: 6
jac: [ 3.725e-09 7.451e-09]
hess_inv: [[ 6.055e+00 -4.237e-01]
[-4.237e-01 3.103e-02]]
nfev: 33
njev: 11
Store the slope and intercept, predict the values directly from the straight line:
inter_ss, slope_ss = mr_ss.x
predicted_ss = inter_ss + slope_ss * hemoglobin
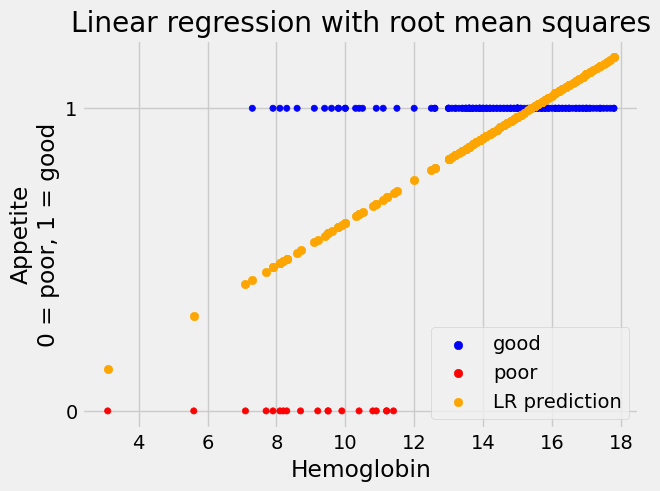
Show the results:
Show code cell content
# Do the base plot of the hemoglobin and appetite_d.
plot_hgb_app()
# A new plot on top of the old.
plt.scatter(hemoglobin, predicted_ss,
label='LR prediction',
color='orange')
plt.title("Linear regression with root mean squares")
# Show the legend.
plt.legend();
Let us remind ourselves of how the root mean squared error values change as we change the slope and the intercept. First we hold the slope constant at a fairly bad guess of 0.1, and try different intercepts. For each intercept we calculate the root mean squared error:
# Some intercepts to try.
intercepts = np.linspace(-2, 1, 1000)
n_inters = len(intercepts)
rmse_errors = np.zeros(n_inters)
for i in np.arange(n_inters):
rmse_errors[i] = rmse_any_line([intercepts[i], 0.1], hemoglobin, appetite_d)
plt.scatter(intercepts, rmse_errors)
plt.xlabel('intercept')
plt.ylabel('Linear SS error')
plt.title("Errors for different intercepts, slope 0.1")
Text(0.5, 1.0, 'Errors for different intercepts, slope 0.1')
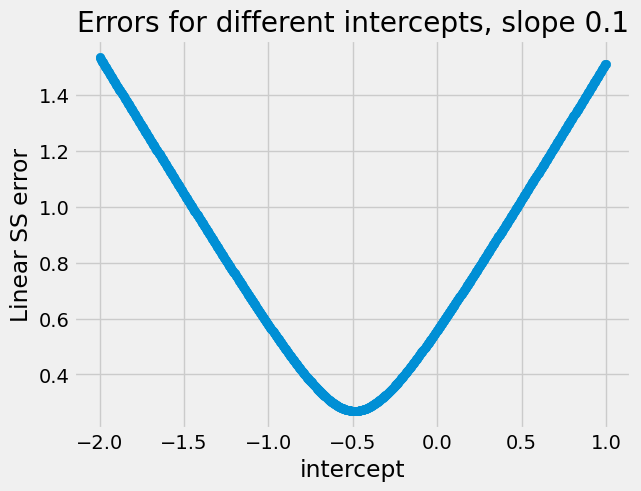
Notice the very simple shape of this curve. It is a V-shape, turning into a
parabola at the bottom. The curve descends steeply for values far from the
minimum, and more slowly when it gets closer. This is a curve that minimize
finds it very easy to work with, because every time it tries an intercept (in
this case), the direction (up, down) of the curve tells minimize what
direction to go next. If the curve is going down at this point, it should try
a larger (more positive) intercept value; if the curve is going up, it should
try a smaller (more negative) intercept. The up/down-ness of the curve tells
minimize the right way to go, and this direction is always correct. You may
also have noticed that this V-shape / parabola form is always the same for
these simple least squares-related functions, like rmse_any_line.
Just to illustrate again, here we try holding the intercept constant at a fairly bad guess of 0.5, and vary the slopes. Notice we get the same helpful V / parabola shape:
# Slopes to try.
slopes = np.linspace(-0.25, 0.25, 1000)
n_slopes = len(slopes)
rmse_errors = np.zeros(n_slopes)
for i in np.arange(n_slopes):
rmse_errors[i] = rmse_any_line([0.5, slopes[i]], hemoglobin, appetite_d)
plt.scatter(slopes, rmse_errors)
plt.xlabel('slope')
plt.ylabel('Linear SS error')
plt.title("Errors for different slopes, intercept 0.5");
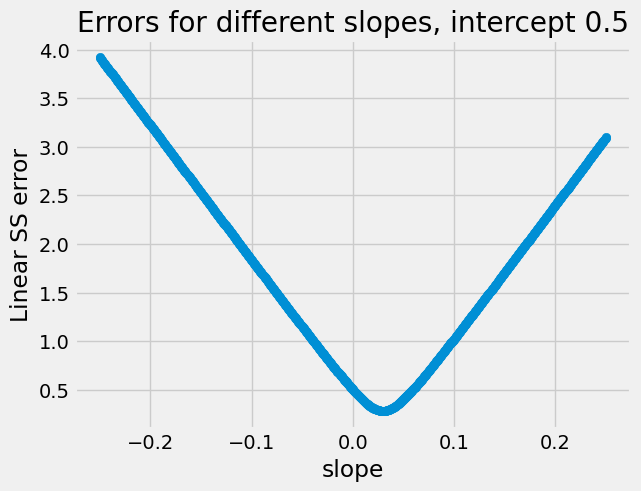
These are plots of how the value of the cost function changes as we change
the parameters. The curves we see above are examples of curves that are
convex; convex curves like
parabolas are particularly easy and quick for minimize to work with.
We will see that using root mean squared error with our preferred sigmoid
prediction generates cost function curves that are a lot more complicated,
making it more difficult for minimize to find the best parameters. If we
give minimize a bad initial guess, it can get the answer badly wrong. Put
technically, this is because the cost function curves are not convex.
Sigmoid prediction with root mean squares error#
For the reasons you saw in the [logistic regression page](logistic regression), we recoil from the very simple straight line fit above, and prefer to use a sigmoid curve to fit the 0 / 1 labels.
In that page we defined the functions to convert the straight line predictions
that we want to use with minimize and the sigmoid predictions:
def inv_logit(y):
""" Reverse logit transformation
"""
odds_ratios = np.exp(y) # Reverse the log operation.
return odds_ratios / (odds_ratios + 1) # Reverse odds ratios operation.
def params2pps(intercept, slope, x):
""" Calculate predicted probabilities of 1 for each observation.
"""
# Predicted log odds of being in class 1.
predicted_log_odds = intercept + slope * x
return inv_logit(predicted_log_odds)
This allowed us to build our root mean square logit cost function. This function calculates the root mean square difference from the sigmoid predictions and the actual 0 / 1 labels.
def rmse_logit_cost(c_s, x_values, y_values):
# Unpack intercept and slope into values.
intercept, slope = c_s
# Predicted p values on sigmoid
pps = params2pps(intercept, slope, x_values)
# Prediction errors.
sigmoid_error = y_values - pps
# Root mean squared error.
return np.sqrt(np.mean(sigmoid_error ** 2))
Then we found our root mean squares best straight line (that corresponds to a
sigmoid after transformation). Notice that we have started minimize with a
guessed intercept of -7 and a guessed slope of 0.8.
mr_rmse_logit = minimize(rmse_logit_cost, [-7, 0.8], args=(hemoglobin, appetite_d))
mr_rmse_logit
message: Optimization terminated successfully.
success: True
status: 0
fun: 0.24677033002934054
x: [-5.281e+00 5.849e-01]
nit: 14
jac: [ 5.774e-08 7.190e-07]
hess_inv: [[ 4.814e+02 -4.559e+01]
[-4.559e+01 4.516e+00]]
nfev: 51
njev: 17
We can calculate the predicted 0 / 1 labels, and plot them.
inter_rmse_logit, slope_rmse_logit = mr_rmse_logit.x
predicted_rmse_logit = params2pps(inter_rmse_logit, slope_rmse_logit, hemoglobin)
Show code cell content
plot_hgb_app()
# A new plot on top of the old.
plt.scatter(hemoglobin, predicted_rmse_logit,
label='Logit ss solution',
color='gold')
# Show the legend.
plt.legend();
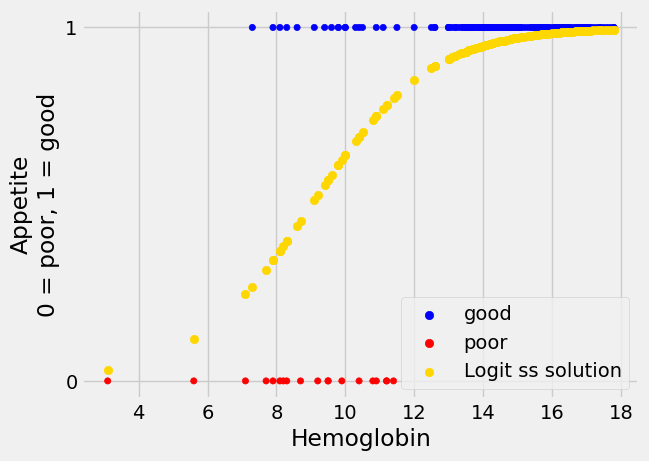
Let us have a look at what the cost function curves look like for the
ss_logit_cost cost function. For now, let us look at what happens to the
cost function curves as we change the intercept, holding the slope the same.
Because we will do this several times, with various intercepts and constant slopes, we make a function, so we don’t repeat ourselves:
def plot_some_intercepts(cost_function, intercepts, slope):
""" Plot values of `cost_function` for given `intercepts` and `slope`
Parameters
----------
cost_function : function
Function to call to get cost function value, given an intercept and a slope.
intercepts : array
Array of intercepts for which to calculate cost function.
slope : number
Slope (held constant for each intercept).
"""
n = len(intercepts)
ss_errors = np.zeros(n)
for i in np.arange(n):
# Calculate and store cost function value for intercept, slope.
ss_errors[i] = cost_function([intercepts[i], slope],
hemoglobin, appetite_d)
plt.plot(intercepts, ss_errors, linewidth=1)
plt.xlabel('intercept')
plt.ylabel('Cost function value');
plt.title('Errors for slope = %.2f' % slope)
Remember the attractive parabolas for the cost function curves in the crude case above, where we were doing simple regression.
In the next cell, we set the slope to the best slope that minimize
found, and show the effect on our ss_logit_cost function, when varying the intercept.
# Cost function curve varying intercept between -20 and 5, for best slope.
plot_some_intercepts(rmse_logit_cost, np.linspace(-20, 5, 1000), slope_rmse_logit)
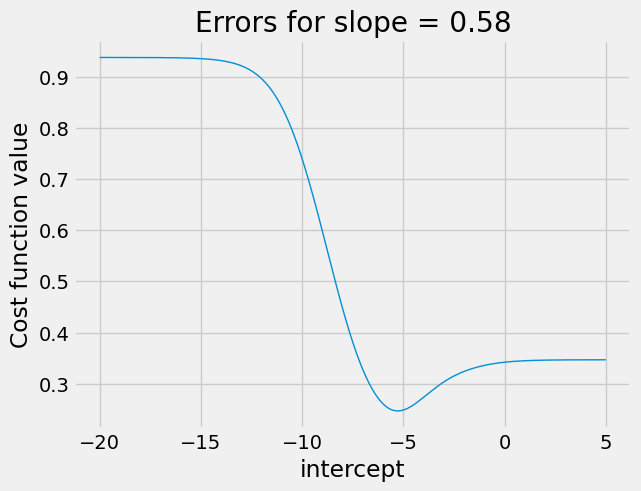
There is a clear minimum at around -5.2, as we expect from the results above,
but we have lost the nice parabola shape. For intercepts greater than about
3, the graph is very flat. This could spell trouble for minimize, if it
gets stuck trying a series of intercepts more than 3. For example, the cost
function will stay almost the same as minimize tries values around 5, so
minimize may not discover that it needs to track back to the real minimum,
near -5.
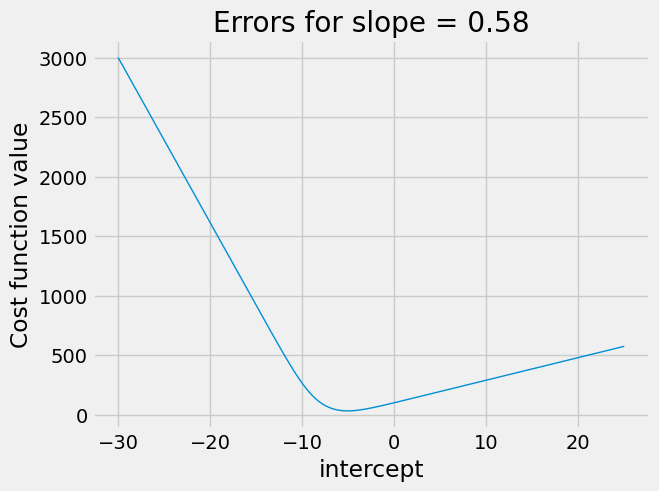
It can get even worse when trying slopes that are further away from the optimum. In the next plot, we set the slope badly wrong, at 3, and try different intercepts.
plot_some_intercepts(rmse_logit_cost, np.linspace(-30, 25, 1000), 3)
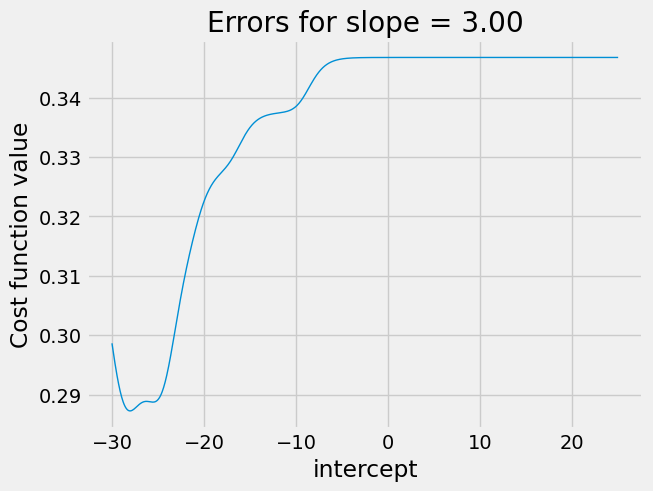
The plot is a strange shape. Again we see a nasty plateau with intercepts
above 0. If minimize is trying intercepts above 0, the cost function may
not vary much, and minimize may get stuck on this plateau, for example
concluding the intercept of 6 is as good as any nearby.
An in fact, this does happen if we set very bad starting estimates for
minimize. Here we set the starting intercept to be 6, and the starting
slope to be 2.5.
bad_mr_rmse_logit = minimize(rmse_logit_cost, [6, 2.5],
args=(hemoglobin, appetite_d))
bad_mr_rmse_logit
message: Optimization terminated successfully.
success: True
status: 0
fun: 0.3467753610259536
x: [ 6.000e+00 2.500e+00]
nit: 0
jac: [ 1.863e-08 5.960e-08]
hess_inv: [[1 0]
[0 1]]
nfev: 3
njev: 1
You can see that minimize has got stuck on the plateau we saw above and has
given up, simply returning the terrible intercept and slope we sent it.
You can also see that minimize did not detect any problems, and returned the
message “Optimization terminated successfully”.
We have this problem because of the irregular shape of the cost-function curve for our cost function, that calculates root mean squared error for the sigmoid predictions.
Back to maximum likelihood#
The logistic_regression page proposed an alternative cost function for the sigmoid predictions - maximum likelihood. See that page for details, but here is somewhat cleaned up version the maximum likelihood cost function from the page above.
def ml_logit_cost(intercept_and_slope, x, y):
""" Cost function for maximum likelihood
"""
intercept, slope = intercept_and_slope
pp1 = params2pps(intercept, slope, x)
p_of_y = y * pp1 + (1 - y) * (1 - pp1)
log_likelihood = np.sum(np.log(p_of_y))
return -log_likelihood
We find the best intercept and slope using the maximum likelihood (ML). While we are at it, we send in the same terrible estimate for the intercept and slope:
mr_ML = minimize(ml_logit_cost, [6, 2.5], args=(hemoglobin, appetite_d))
mr_ML
message: Optimization terminated successfully.
success: True
status: 0
fun: 29.16799389586808
x: [-7.292e+00 7.992e-01]
nit: 12
jac: [-2.623e-06 9.060e-06]
hess_inv: [[ 2.436e+00 -2.276e-01]
[-2.276e-01 2.231e-02]]
nfev: 87
njev: 29
The fit is reasonable:
inter_ML, slope_ML = mr_ML.x
predicted_ML = inv_logit(inter_ML + slope_ML * hemoglobin)
Show code cell content
plot_hgb_app()
plt.scatter(hemoglobin, predicted_ML, c='gold', label='ML prediction')
plt.legend();
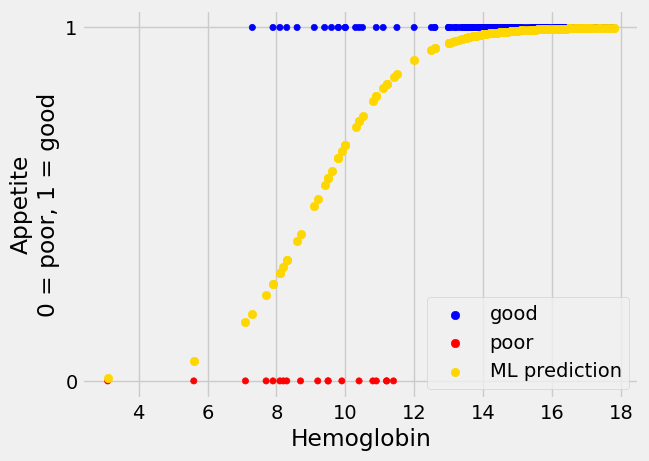
The ML search by minimize is more reliable than the sum-of-squares case
above; it is less dependent on us choosing some reasonable starting values.
This is because the ML cost function is convex. Here is the cost-function
curve for the ML cost function, as we vary the intercept for a fixed slope. We
see that we have a much more predictable curve, that slopes smoothly downwards
to a minimum and smoothly upwards after that.
plot_some_intercepts(ml_logit_cost, np.linspace(-30, 25, 1000), slope_rmse_logit)
However, this does not mean the ML cost function is infallible. We can push it to a state where the calculation errors start to overwhelm the values. However, ML still has the advantage, because, unlike the root mean square, we do get a warning:
# Starting estimates too far off.
minimize(ml_logit_cost, [6, 3], args=(hemoglobin, appetite_d))
/opt/hostedtoolcache/Python/3.11.9/x64/lib/python3.11/site-packages/pandas/core/arraylike.py:399: RuntimeWarning: divide by zero encountered in log
result = getattr(ufunc, method)(*inputs, **kwargs)
/opt/hostedtoolcache/Python/3.11.9/x64/lib/python3.11/site-packages/pandas/core/arraylike.py:399: RuntimeWarning: divide by zero encountered in log
result = getattr(ufunc, method)(*inputs, **kwargs)
/opt/hostedtoolcache/Python/3.11.9/x64/lib/python3.11/site-packages/scipy/optimize/_numdiff.py:590: RuntimeWarning: invalid value encountered in subtract
df = fun(x) - f0
/opt/hostedtoolcache/Python/3.11.9/x64/lib/python3.11/site-packages/pandas/core/arraylike.py:399: RuntimeWarning: divide by zero encountered in log
result = getattr(ufunc, method)(*inputs, **kwargs)
/opt/hostedtoolcache/Python/3.11.9/x64/lib/python3.11/site-packages/scipy/optimize/_numdiff.py:590: RuntimeWarning: invalid value encountered in subtract
df = fun(x) - f0
message: NaN result encountered.
success: False
status: 3
fun: inf
x: [ 6.000e+00 3.000e+00]
nit: 0
jac: [ nan nan]
hess_inv: [[1 0]
[0 1]]
nfev: 3
njev: 1
Another example#
You will find another demonstration of the difference between root mean square and maximum likelihood in this page on logistic cost functions.