Statsmodels where columns have spaces#
Show code cell content
import numpy as np
import pandas as pd
# Safe setting for Pandas. Needs Pandas version >= 1.5.
pd.set_option('mode.copy_on_write', True)
import matplotlib.pyplot as plt
plt.style.use('fivethirtyeight')
np.set_printoptions(suppress=True)
import statsmodels.formula.api as smf
Back at the simple and multiple regression page, we introduced the StatsModels library.
As you may remember from that page, we were looking for the best least-squares straight-line fit between two columns of values in patients with chronic kidney disease:
ckd = pd.read_csv('ckd.csv')
ckd.head()
| Age | Blood Pressure | Specific Gravity | Albumin | Sugar | Red Blood Cells | Pus Cell | Pus Cell clumps | Bacteria | Blood Glucose Random | ... | Packed Cell Volume | White Blood Cell Count | Red Blood Cell Count | Hypertension | Diabetes Mellitus | Coronary Artery Disease | Appetite | Pedal Edema | Anemia | Class | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 48 | 70 | 1.005 | 4 | 0 | normal | abnormal | present | notpresent | 117 | ... | 32 | 6700 | 3.9 | yes | no | no | poor | yes | yes | 1 |
| 1 | 53 | 90 | 1.020 | 2 | 0 | abnormal | abnormal | present | notpresent | 70 | ... | 29 | 12100 | 3.7 | yes | yes | no | poor | no | yes | 1 |
| 2 | 63 | 70 | 1.010 | 3 | 0 | abnormal | abnormal | present | notpresent | 380 | ... | 32 | 4500 | 3.8 | yes | yes | no | poor | yes | no | 1 |
| 3 | 68 | 80 | 1.010 | 3 | 2 | normal | abnormal | present | present | 157 | ... | 16 | 11000 | 2.6 | yes | yes | yes | poor | yes | no | 1 |
| 4 | 61 | 80 | 1.015 | 2 | 0 | abnormal | abnormal | notpresent | notpresent | 173 | ... | 24 | 9200 | 3.2 | yes | yes | yes | poor | yes | yes | 1 |
5 rows × 25 columns
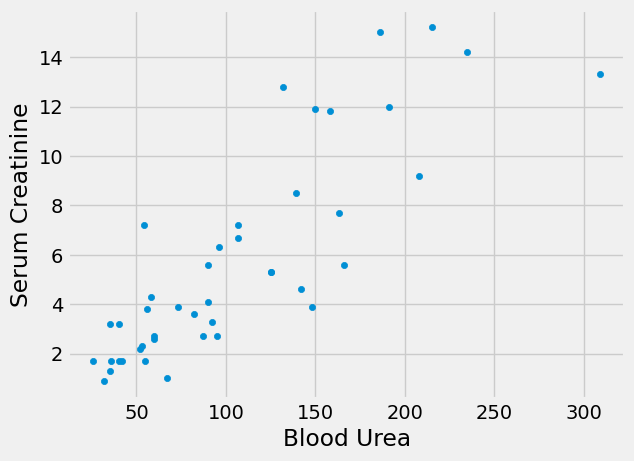
In particular we were interested to predict the blood concentration of creatinine (a marker of kidney failure) from the blood concentration of urea (another marker of kidney failure), in patients with chronic kidney disease.
Notice that creatinine and urea columns are "Serum Creatinine"
and "Blood Urea" respectively. Both columns have spaces in their names.
We first select our columns of interest, and restrict ourselves to rows for patients with chronic kidney disease (CKD):
# Data frame restricted to kidney patients and columns of interest.
ckdp = ckd.loc[
ckd['Class'] == 1, # rows for CKD
['Serum Creatinine', 'Blood Urea']] # columns of interest
ckdp.head()
| Serum Creatinine | Blood Urea | |
|---|---|---|
| 0 | 3.8 | 56 |
| 1 | 7.2 | 107 |
| 2 | 2.7 | 60 |
| 3 | 4.1 | 90 |
| 4 | 3.9 | 148 |
ckdp.plot.scatter('Blood Urea', 'Serum Creatinine')
<Axes: xlabel='Blood Urea', ylabel='Serum Creatinine'>
In the single and multiple regression page, we started by renaming the columns, where the new names did not have spaces.
ckdp_renamed = ckdp.copy()
# Rename the columns to names without spaces.
ckdp_renamed.columns = ['Creatinine', 'Urea']
ckdp_renamed.head()
| Creatinine | Urea | |
|---|---|---|
| 0 | 3.8 | 56 |
| 1 | 7.2 | 107 |
| 2 | 2.7 | 60 |
| 3 | 4.1 | 90 |
| 4 | 3.9 | 148 |
We then ran a simple regression model in Statsmodels, to find the least-squares straight line.
simple_model = smf.ols(formula="Creatinine ~ Urea", data=ckdp_renamed)
simple_fit = simple_model.fit()
simple_fit.summary()
| Dep. Variable: | Creatinine | R-squared: | 0.716 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.709 |
| Method: | Least Squares | F-statistic: | 103.1 |
| Date: | Wed, 05 Jun 2024 | Prob (F-statistic): | 9.33e-13 |
| Time: | 16:06:01 | Log-Likelihood: | -95.343 |
| No. Observations: | 43 | AIC: | 194.7 |
| Df Residuals: | 41 | BIC: | 198.2 |
| Df Model: | 1 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | -0.0849 | 0.668 | -0.127 | 0.899 | -1.434 | 1.264 |
| Urea | 0.0552 | 0.005 | 10.155 | 0.000 | 0.044 | 0.066 |
| Omnibus: | 2.409 | Durbin-Watson: | 1.303 |
|---|---|---|---|
| Prob(Omnibus): | 0.300 | Jarque-Bera (JB): | 1.814 |
| Skew: | 0.503 | Prob(JB): | 0.404 |
| Kurtosis: | 3.043 | Cond. No. | 236. |
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
But, if we wanted to use the original column names, we would have to do some extra work to make Statsmodels accept column names with spaces. And in fact we have to do the same thing if there are special characters, which, like the spaces would make the column names invalid as variable names.
For example, let’s say we were using the DataFrame ckdp with the original
column names. We could try this:
# This generates an error, because the Statsmodels formula interface
# needs column names that work as variable names.
another_model = smf.ols(formula="Serum Creatinine ~ Blood Urea",
data=ckdp)
Traceback (most recent call last):
File /opt/hostedtoolcache/Python/3.11.9/x64/lib/python3.11/site-packages/IPython/core/interactiveshell.py:3577 in run_code
exec(code_obj, self.user_global_ns, self.user_ns)
Cell In[7], line 3
another_model = smf.ols(formula="Serum Creatinine ~ Blood Urea",
File /opt/hostedtoolcache/Python/3.11.9/x64/lib/python3.11/site-packages/statsmodels/base/model.py:203 in from_formula
tmp = handle_formula_data(data, None, formula, depth=eval_env,
File /opt/hostedtoolcache/Python/3.11.9/x64/lib/python3.11/site-packages/statsmodels/formula/formulatools.py:63 in handle_formula_data
result = dmatrices(formula, Y, depth, return_type='dataframe',
File /opt/hostedtoolcache/Python/3.11.9/x64/lib/python3.11/site-packages/patsy/highlevel.py:309 in dmatrices
(lhs, rhs) = _do_highlevel_design(formula_like, data, eval_env,
File /opt/hostedtoolcache/Python/3.11.9/x64/lib/python3.11/site-packages/patsy/highlevel.py:164 in _do_highlevel_design
design_infos = _try_incr_builders(formula_like, data_iter_maker, eval_env,
File /opt/hostedtoolcache/Python/3.11.9/x64/lib/python3.11/site-packages/patsy/highlevel.py:66 in _try_incr_builders
return design_matrix_builders([formula_like.lhs_termlist,
File /opt/hostedtoolcache/Python/3.11.9/x64/lib/python3.11/site-packages/patsy/build.py:689 in design_matrix_builders
factor_states = _factors_memorize(all_factors, data_iter_maker, eval_env)
File /opt/hostedtoolcache/Python/3.11.9/x64/lib/python3.11/site-packages/patsy/build.py:354 in _factors_memorize
which_pass = factor.memorize_passes_needed(state, eval_env)
File /opt/hostedtoolcache/Python/3.11.9/x64/lib/python3.11/site-packages/patsy/eval.py:478 in memorize_passes_needed
subset_names = [name for name in ast_names(self.code)
File /opt/hostedtoolcache/Python/3.11.9/x64/lib/python3.11/site-packages/patsy/eval.py:478 in <listcomp>
subset_names = [name for name in ast_names(self.code)
File /opt/hostedtoolcache/Python/3.11.9/x64/lib/python3.11/site-packages/patsy/eval.py:109 in ast_names
for node in ast.walk(ast.parse(code)):
File /opt/hostedtoolcache/Python/3.11.9/x64/lib/python3.11/ast.py:50 in parse
return compile(source, filename, mode, flags,
File <unknown>:1
Serum Creatinine
^
SyntaxError: invalid syntax
The solution is to use the Q()
(Quote)
function in your formula. It tells Statsmodels that you mean the words ‘Serum’
and ‘Creatinine’ to be one thing: ‘Serum Creatinine’ - the name of the column.
another_model = smf.ols(formula="Q('Serum Creatinine') ~ Q('Blood Urea')",
data=ckdp)
another_fit = another_model.fit()
another_fit.summary()
| Dep. Variable: | Q('Serum Creatinine') | R-squared: | 0.716 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.709 |
| Method: | Least Squares | F-statistic: | 103.1 |
| Date: | Wed, 05 Jun 2024 | Prob (F-statistic): | 9.33e-13 |
| Time: | 16:06:01 | Log-Likelihood: | -95.343 |
| No. Observations: | 43 | AIC: | 194.7 |
| Df Residuals: | 41 | BIC: | 198.2 |
| Df Model: | 1 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | -0.0849 | 0.668 | -0.127 | 0.899 | -1.434 | 1.264 |
| Q('Blood Urea') | 0.0552 | 0.005 | 10.155 | 0.000 | 0.044 | 0.066 |
| Omnibus: | 2.409 | Durbin-Watson: | 1.303 |
|---|---|---|---|
| Prob(Omnibus): | 0.300 | Jarque-Bera (JB): | 1.814 |
| Skew: | 0.503 | Prob(JB): | 0.404 |
| Kurtosis: | 3.043 | Cond. No. | 236. |
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.