Dimension reduction with Principal Component Analysis#
See the PCA introduction for background.
Start by loading the libraries we need, and doing some configuration:
import numpy as np
import numpy.linalg as npl
# Display array values to 6 digits of precision
np.set_printoptions(precision=6, suppress=True)
import pandas as pd
pd.set_option('mode.copy_on_write', True)
import matplotlib.pyplot as plt
from sklearn.decomposition import PCA
from sklearn.linear_model import LinearRegression
from sklearn.metrics import r2_score
As for the PCA page above, we will use the English Premier League (EPL) table data for this example. See the EPL dataset page for more on this dataset.
df = pd.read_csv('data/premier_league_2021.csv')
df.head()
| rank | team | played | won | drawn | lost | for | against | goal_difference | points | wages_year | keeper | defense | midfield | forward | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | Manchester City | 38 | 29 | 6 | 3 | 99 | 26 | 73 | 93 | 168572 | 8892 | 60320 | 24500 | 74860 |
| 1 | 2 | Liverpool | 38 | 28 | 8 | 2 | 94 | 26 | 68 | 92 | 148772 | 14560 | 46540 | 47320 | 40352 |
| 2 | 3 | Chelsea | 38 | 21 | 11 | 6 | 76 | 33 | 43 | 74 | 187340 | 12480 | 51220 | 51100 | 72540 |
| 3 | 4 | Tottenham Hotspur | 38 | 22 | 5 | 11 | 69 | 40 | 29 | 71 | 110416 | 6760 | 29516 | 30680 | 43460 |
| 4 | 5 | Arsenal | 38 | 22 | 3 | 13 | 61 | 48 | 13 | 69 | 118074 | 8400 | 37024 | 27300 | 45350 |
We want to predict goal difference from spending on the various position types.
We have spending data on keeper defense, midfield and forward.
y = df['goal_difference']
# Divide spending by 10,000 GBP to make numbers easier to read.
X = df[['keeper', 'defense', 'midfield', 'forward']] / 10_000
X.head()
| keeper | defense | midfield | forward | |
|---|---|---|---|---|
| 0 | 0.8892 | 6.0320 | 2.450 | 7.4860 |
| 1 | 1.4560 | 4.6540 | 4.732 | 4.0352 |
| 2 | 1.2480 | 5.1220 | 5.110 | 7.2540 |
| 3 | 0.6760 | 2.9516 | 3.068 | 4.3460 |
| 4 | 0.8400 | 3.7024 | 2.730 | 4.5350 |
basic_reg = LinearRegression().fit(X, y)
basic_reg.coef_
array([-15.502165, 24.888585, 6.337194, -5.453179])
How good is the model fit?
fitted = basic_reg.predict(X)
fitted
array([ 58.702936, 48.899144, 48.61422 , 6.380567, 19.351939,
15.741829, -7.189155, -0.542271, -14.657475, -10.357978,
5.580663, -11.703874, -33.987123, -10.908234, -38.285969,
12.001697, -30.212118, -22.902239, -14.680438, -19.84612 ])
full_r2 = r2_score(y, fitted)
full_r2
0.6593261703105764
It turns out the variables (features) are highly correlated, one with another.
X.corr()
| keeper | defense | midfield | forward | |
|---|---|---|---|---|
| keeper | 1.000000 | 0.770622 | 0.653607 | 0.836858 |
| defense | 0.770622 | 1.000000 | 0.792146 | 0.918498 |
| midfield | 0.653607 | 0.792146 | 1.000000 | 0.718424 |
| forward | 0.836858 | 0.918498 | 0.718424 | 1.000000 |
We want to reduce these four variables down to fewer variables, while still capturing the same information.
Let us first run PCA on the four columns:
pca = PCA()
pca = pca.fit(X)
# Scikit-learn's components.
pca.components_
array([[ 0.162258, 0.467464, 0.330791, 0.803571],
[-0.044097, 0.155829, 0.881052, -0.444433],
[-0.229876, 0.859756, -0.324967, -0.319959],
[ 0.958585, 0.134218, -0.093391, -0.233192]])
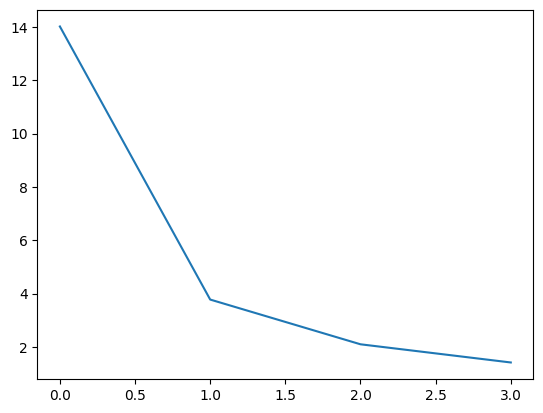
Review the scree-plot of the component weights:
plt.plot(pca.singular_values_)
[<matplotlib.lines.Line2D at 0x7f6b17deb110>]
We can calculate the amount of each row explained by each component, by using fit_transform of the PCA object:
comp_vals = pca.fit_transform(X)
comp_vals
array([[ 4.885102, -0.949893, 1.316577, -0.44375 ],
[ 2.314803, 2.354588, 0.364081, 0.506204],
[ 5.211402, 1.339186, -0.33846 , -0.416269],
[ 1.091745, 0.5195 , -0.478962, -0.387058],
[ 1.509396, 0.247471, 0.178209, -0.141586],
[ 8.670343, -1.625328, -0.7396 , 0.447318],
[-0.853819, -0.589592, 0.539047, 0.666987],
[-0.789195, 0.171897, 0.305088, 0.445527],
[-2.274588, -0.086226, 0.035262, -0.101522],
[-1.31331 , -0.407973, -0.022812, -0.397283],
[-1.053835, 0.15501 , 0.434515, -0.011451],
[-1.112012, 0.134205, -0.260818, -0.058728],
[-3.420161, -0.259338, -0.349634, -0.116371],
[-0.140709, -0.665132, -0.05001 , 0.032668],
[-1.937029, -0.105797, -0.84301 , 0.271394],
[ 0.893833, 0.785698, -0.225254, -0.144854],
[-2.666776, -0.942176, -0.100787, -0.215631],
[-2.68459 , -0.439447, 0.004598, -0.078675],
[-2.915953, 0.472992, -0.047468, -0.043406],
[-3.414648, -0.109644, 0.279437, 0.186487]])
If you have followed the PCA introduction page, you will know you can also get these scores by matrix multiplication:
# Getting component values by using matrix multiplication.
demeaned_X = X - np.mean(X, axis=0)
demeaned_X.dot(pca.components_.T)
| 0 | 1 | 2 | 3 | |
|---|---|---|---|---|
| 0 | 4.885102 | -0.949893 | 1.316577 | -0.443750 |
| 1 | 2.314803 | 2.354588 | 0.364081 | 0.506204 |
| 2 | 5.211402 | 1.339186 | -0.338460 | -0.416269 |
| 3 | 1.091745 | 0.519500 | -0.478962 | -0.387058 |
| 4 | 1.509396 | 0.247471 | 0.178209 | -0.141586 |
| 5 | 8.670343 | -1.625328 | -0.739600 | 0.447318 |
| 6 | -0.853819 | -0.589592 | 0.539047 | 0.666987 |
| 7 | -0.789195 | 0.171897 | 0.305088 | 0.445527 |
| 8 | -2.274588 | -0.086226 | 0.035262 | -0.101522 |
| 9 | -1.313310 | -0.407973 | -0.022812 | -0.397283 |
| 10 | -1.053835 | 0.155010 | 0.434515 | -0.011451 |
| 11 | -1.112012 | 0.134205 | -0.260818 | -0.058728 |
| 12 | -3.420161 | -0.259338 | -0.349634 | -0.116371 |
| 13 | -0.140709 | -0.665132 | -0.050010 | 0.032668 |
| 14 | -1.937029 | -0.105797 | -0.843010 | 0.271394 |
| 15 | 0.893833 | 0.785698 | -0.225254 | -0.144854 |
| 16 | -2.666776 | -0.942176 | -0.100787 | -0.215631 |
| 17 | -2.684590 | -0.439447 | 0.004598 | -0.078675 |
| 18 | -2.915953 | 0.472992 | -0.047468 | -0.043406 |
| 19 | -3.414648 | -0.109644 | 0.279437 | 0.186487 |
We can use these four columns instead of the original four columns as regressors. Think of the component scores (values) as being a re-expression of the original four columns, that keeps all the same information. When we fit this model, the component scores will get their own coefficients, but because those columns contain the same information as the original columns, the resulting fitted values will be the same as for the original model (within the precision of the calculations).
comp_reg = LinearRegression().fit(comp_vals, y)
comp_fitted = comp_reg.predict(comp_vals)
# The columns and parameters are different, but the fitted values
# are the same.
np.allclose(comp_fitted, fitted)
True
Now we’ve seen the scree-plot, we might conclude that only the first component is important, and we can discard the others. We then just include the first component values a new model.
comp1_vals = comp_vals[:, :1] # A 2D array with one column
comp1_vals
array([[ 4.885102],
[ 2.314803],
[ 5.211402],
[ 1.091745],
[ 1.509396],
[ 8.670343],
[-0.853819],
[-0.789195],
[-2.274588],
[-1.31331 ],
[-1.053835],
[-1.112012],
[-3.420161],
[-0.140709],
[-1.937029],
[ 0.893833],
[-2.666776],
[-2.68459 ],
[-2.915953],
[-3.414648]])
comp1_reg = LinearRegression().fit(comp1_vals, y)
comp1_reg.coef_
array([6.833448])
We don’t have quite the same quality of model fit — as measured by \(R^2\), for example, but we have recovered most of the \(R^2\) using this single regressor instead of the original four.
comp1_fitted = comp1_reg.predict(comp1_vals)
r2_score(y, comp1_fitted)
0.42171885321184366
We could improve the fit a little by adding the second component:
comp12_reg = LinearRegression().fit(comp_vals[:, :2], y)
r2_score(y, comp12_reg.predict(comp_vals[:, :2]))
0.525326169266535
And a little more with the third:
comp123_reg = LinearRegression().fit(comp_vals[:, :3], y)
r2_score(y, comp123_reg.predict(comp_vals[:, :3]))
0.6484639642971903
Until we get the full \(R^2\) with all four components (which gives the same underling model as using the original four regressors):
comp_reg = LinearRegression().fit(comp_vals, y)
r2_score(y, comp_reg.predict(comp_vals))
0.6593261703105762