Naive Bayes classifiers#
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
pd.set_option('mode.copy_on_write', True)
Also see: Naive Bayes classifiers in the Python Data Science Handbook.
import seaborn as sns
penguins = pd.read_csv('data/penguins.csv').dropna()
penguins
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | year | |
|---|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | male | 2007 |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | female | 2007 |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | female | 2007 |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | female | 2007 |
| 5 | Adelie | Torgersen | 39.3 | 20.6 | 190.0 | 3650.0 | male | 2007 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 339 | Chinstrap | Dream | 55.8 | 19.8 | 207.0 | 4000.0 | male | 2009 |
| 340 | Chinstrap | Dream | 43.5 | 18.1 | 202.0 | 3400.0 | female | 2009 |
| 341 | Chinstrap | Dream | 49.6 | 18.2 | 193.0 | 3775.0 | male | 2009 |
| 342 | Chinstrap | Dream | 50.8 | 19.0 | 210.0 | 4100.0 | male | 2009 |
| 343 | Chinstrap | Dream | 50.2 | 18.7 | 198.0 | 3775.0 | female | 2009 |
333 rows × 8 columns
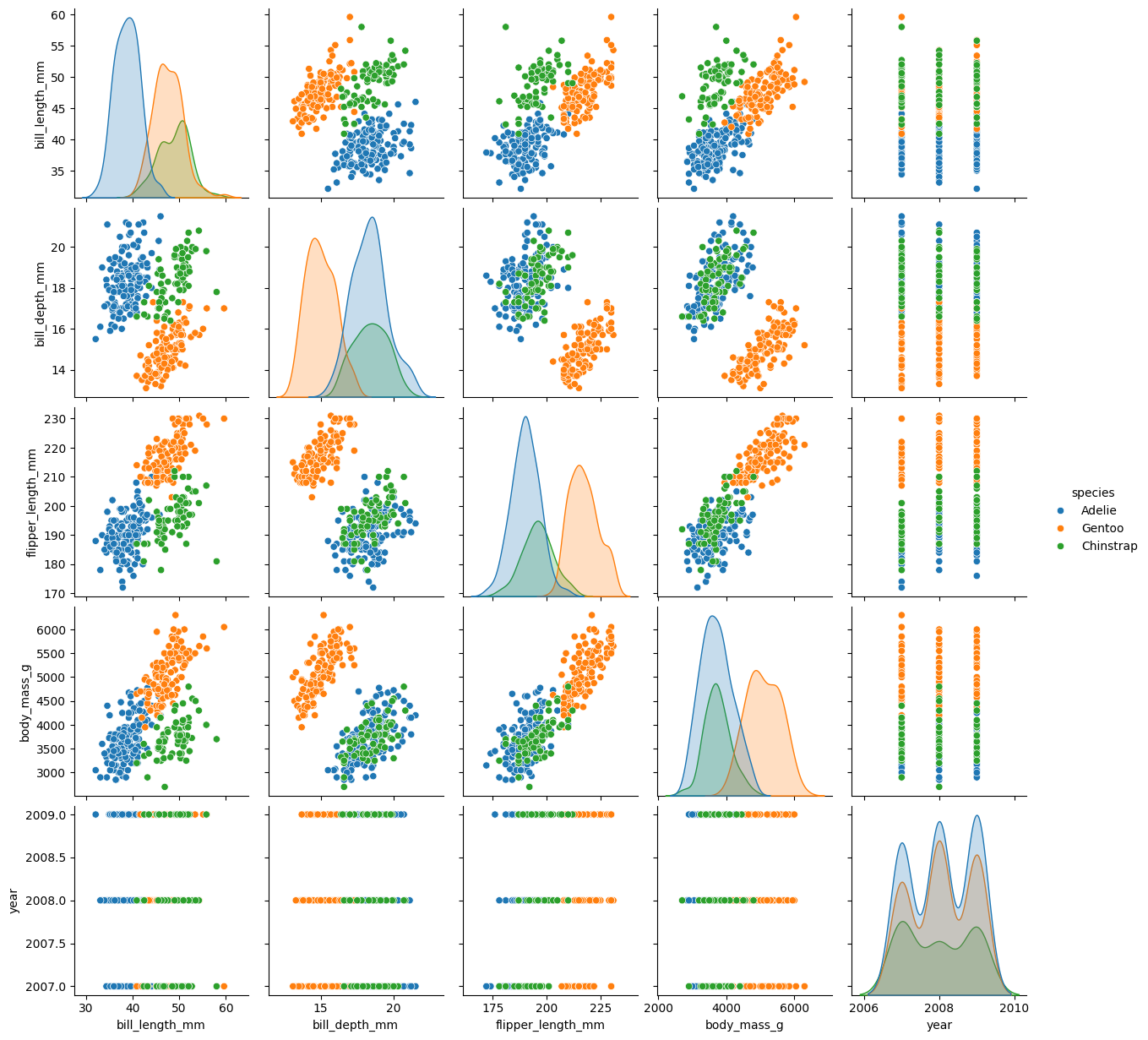
sns.pairplot(penguins, hue="species")
<seaborn.axisgrid.PairGrid at 0x7f5ef1664910>
species_names = ['Chinstrap', 'Gentoo']
df = penguins.loc[
penguins['species'].isin(species_names),
['species', 'body_mass_g', 'bill_depth_mm']
]
df
| species | body_mass_g | bill_depth_mm | |
|---|---|---|---|
| 152 | Gentoo | 4500.0 | 13.2 |
| 153 | Gentoo | 5700.0 | 16.3 |
| 154 | Gentoo | 4450.0 | 14.1 |
| 155 | Gentoo | 5700.0 | 15.2 |
| 156 | Gentoo | 5400.0 | 14.5 |
| ... | ... | ... | ... |
| 339 | Chinstrap | 4000.0 | 19.8 |
| 340 | Chinstrap | 3400.0 | 18.1 |
| 341 | Chinstrap | 3775.0 | 18.2 |
| 342 | Chinstrap | 4100.0 | 19.0 |
| 343 | Chinstrap | 3775.0 | 18.7 |
187 rows × 3 columns
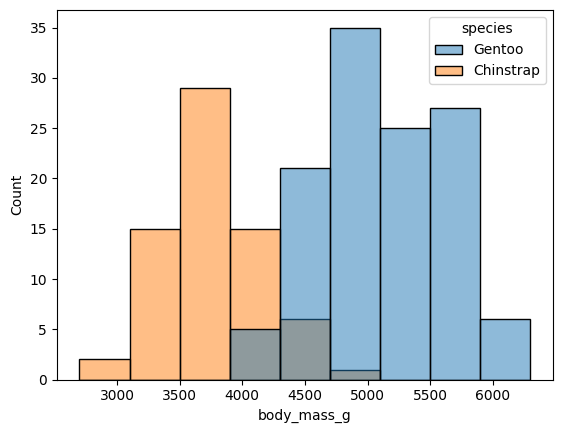
sns.histplot(data=df, x="body_mass_g", hue="species")
<Axes: xlabel='body_mass_g', ylabel='Count'>
by_species = df.groupby('species')
bm_by_species = by_species['body_mass_g']
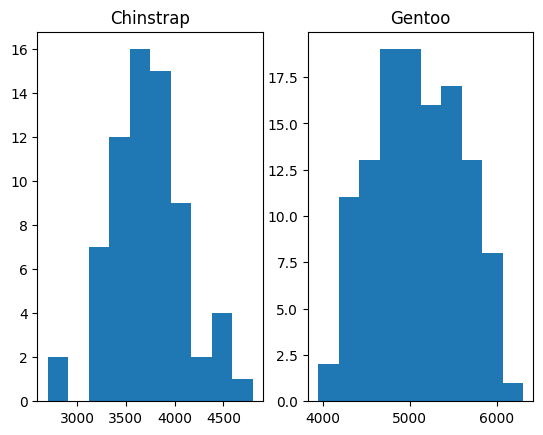
fig, axes = plt.subplots(1, 2)
axes[0].hist(bm_by_species.get_group('Chinstrap'))
axes[0].set_title('Chinstrap')
axes[1].hist(bm_by_species.get_group('Gentoo'));
axes[1].set_title('Gentoo')
Text(0.5, 1.0, 'Gentoo')
Enumerate#
While we are here — we introduce enumerate.
Imagine we need both the elements in a list, and the position of that element:
position_no = 0 # A variable to keep track of the position (index).
for name in species_names:
print('Group', position_no, 'name is', name)
position_no = position_no + 1 # We have to increment the position.
Group 0 name is Chinstrap
Group 1 name is Gentoo
We can avoid the extra variable by using enumerate, wrapped around the set of
things we want to process. enumerate returns two things at each step — the
position (index), and the next element from the set of things.
for position_no, name in enumerate(species_names):
print('Group', position_no, 'name is', name)
Group 0 name is Chinstrap
Group 1 name is Gentoo
We use enumerate to do the plots more neatly below.
The plots again:
fig, axes = plt.subplots(1, 2)
for i, name in enumerate(species_names):
axes[i].hist(bm_by_species.get_group(name))
axes[i].set_title(name)

Towards classification with probability#
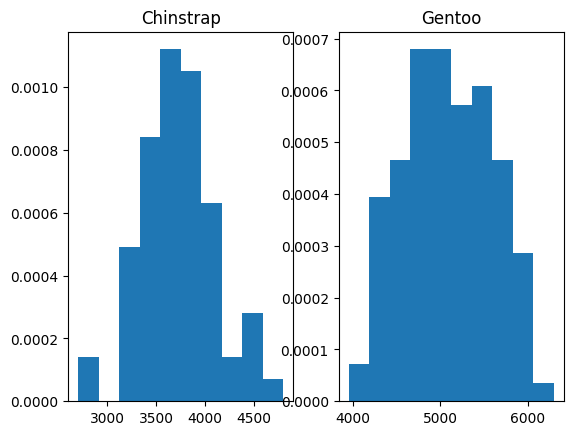
We are going to start to think about getting the probability of a particular mass given we have a Chinstrap or a Gentoo penguin. We might like to start with density histograms. Here was ask Matplotlib to set the area of the bars to sum to 1.
fig, axes = plt.subplots(1, 2)
for i, name in enumerate(species_names):
axes[i].hist(bm_by_species.get_group(name), density=True)
axes[i].set_title(name)
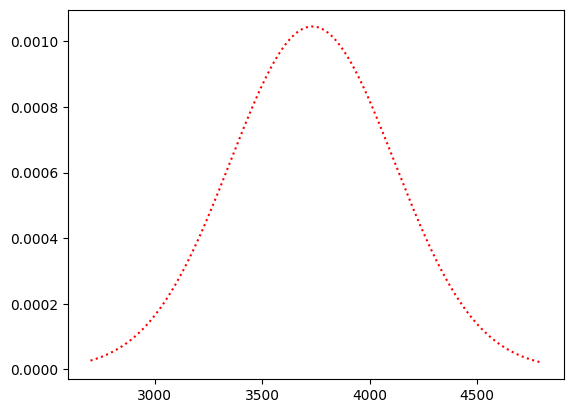
For the moment, let us model these two distributions as normal curves.
# Notice the Numpy std function (variance divisor is n rather than
# (n - 1). This is to match scikit-learn's implementation.
bm_params = bm_by_species.agg(mean="mean", std=lambda x : np.std(x))
bm_params
| mean | std | |
|---|---|---|
| species | ||
| Chinstrap | 3733.088235 | 381.498621 |
| Gentoo | 5092.436975 | 499.364666 |
We can use a Scipy statistics distribution to make a normal distribution with the same mean and standard deviation.
import scipy.stats as sps
# A normal distribution with the same mean and standard deviation.
chinstrap_dist = sps.norm(bm_params.loc['Chinstrap', 'mean'],
bm_params.loc['Chinstrap', 'std'])
# The probability density function of the distribution:
chinstraps = bm_by_species.get_group('Chinstrap')
c_masses_x = np.linspace(chinstraps.min(), chinstraps.max(), 1000)
plt.plot(c_masses_x, chinstrap_dist.pdf(c_masses_x), 'r:')
[<matplotlib.lines.Line2D at 0x7f5ed9acd7d0>]
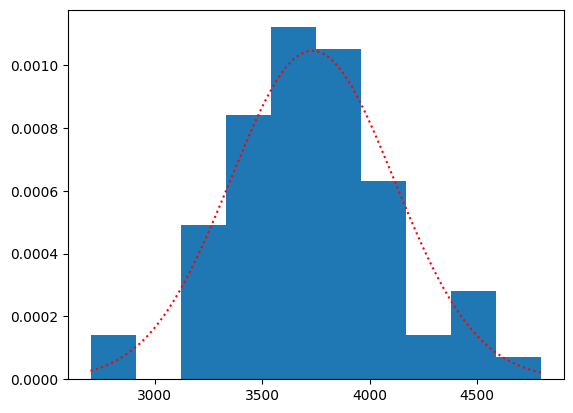
This is the plot along with the actual densities:
plt.hist(chinstraps, density=True)
plt.plot(c_masses_x, chinstrap_dist.pdf(c_masses_x), 'r:')
[<matplotlib.lines.Line2D at 0x7f5ed8647fd0>]
The normal (Gaussian) probability density function gives our estimate of the probability (density) of any given weight.
chinstrap_dist.pdf(3500)
np.float64(0.0008676740928442561)
With these we can get the probability of any mass in the dataset given that the penguin is a Chinstrap:
Actually, to be concise, let’s use \(m\) for \(\text{mass}\) and \(C\) for \(\text{Chinstrap}\).
We fill in the values of \(P(m | C)\):
df['p_m_given_c'] = chinstrap_dist.pdf(df['body_mass_g'])
df
| species | body_mass_g | bill_depth_mm | p_m_given_c | |
|---|---|---|---|---|
| 152 | Gentoo | 4500.0 | 13.2 | 1.386413e-04 |
| 153 | Gentoo | 5700.0 | 16.3 | 1.767096e-09 |
| 154 | Gentoo | 4450.0 | 14.1 | 1.788899e-04 |
| 155 | Gentoo | 5700.0 | 15.2 | 1.767096e-09 |
| 156 | Gentoo | 5400.0 | 14.5 | 7.477526e-08 |
| ... | ... | ... | ... | ... |
| 339 | Chinstrap | 4000.0 | 19.8 | 8.186991e-04 |
| 340 | Chinstrap | 3400.0 | 18.1 | 7.143042e-04 |
| 341 | Chinstrap | 3775.0 | 18.2 | 1.039432e-03 |
| 342 | Chinstrap | 4100.0 | 19.0 | 6.585033e-04 |
| 343 | Chinstrap | 3775.0 | 18.7 | 1.039432e-03 |
187 rows × 4 columns
Likewise:
which we will write as:
gentoo_dist = sps.norm(bm_params.loc['Gentoo', 'mean'],
bm_params.loc['Gentoo', 'std'])
gentoos = bm_by_species.get_group('Gentoo')
g_masses_x = np.linspace(gentoos.min(), gentoos.max(), 1000)
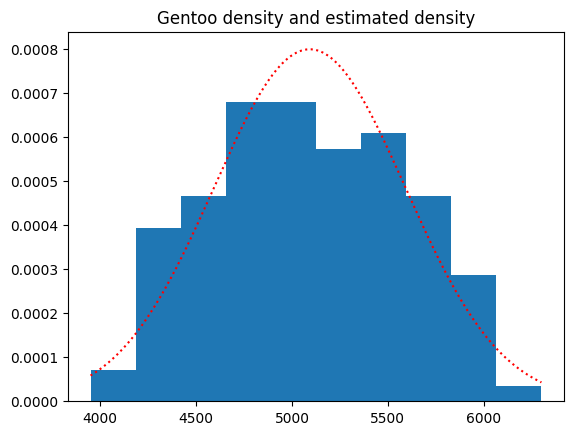
plt.hist(gentoos, density=True)
plt.plot(g_masses_x, gentoo_dist.pdf(g_masses_x), 'r:');
plt.title('Gentoo density and estimated density');
df['p_m_given_g'] = gentoo_dist.pdf(df['body_mass_g'])
df
| species | body_mass_g | bill_depth_mm | p_m_given_c | p_m_given_g | |
|---|---|---|---|---|---|
| 152 | Gentoo | 4500.0 | 13.2 | 1.386413e-04 | 0.000395 |
| 153 | Gentoo | 5700.0 | 16.3 | 1.767096e-09 | 0.000381 |
| 154 | Gentoo | 4450.0 | 14.1 | 1.788899e-04 | 0.000349 |
| 155 | Gentoo | 5700.0 | 15.2 | 1.767096e-09 | 0.000381 |
| 156 | Gentoo | 5400.0 | 14.5 | 7.477526e-08 | 0.000661 |
| ... | ... | ... | ... | ... | ... |
| 339 | Chinstrap | 4000.0 | 19.8 | 8.186991e-04 | 0.000073 |
| 340 | Chinstrap | 3400.0 | 18.1 | 7.143042e-04 | 0.000003 |
| 341 | Chinstrap | 3775.0 | 18.2 | 1.039432e-03 | 0.000025 |
| 342 | Chinstrap | 4100.0 | 19.0 | 6.585033e-04 | 0.000111 |
| 343 | Chinstrap | 3775.0 | 18.7 | 1.039432e-03 | 0.000025 |
187 rows × 5 columns
We also need prior probabilities for Chinstrap (\(C\)) and Gentoo (\(G\)):
Let’s just use the proportions in the dataset:
p_chinstrap = np.mean(df['species'] == 'Chinstrap')
p_chinstrap
np.float64(0.36363636363636365)
p_gentoo = np.mean(df['species'] == 'Gentoo')
p_gentoo
np.float64(0.6363636363636364)
df['p_chinstrap'] = p_chinstrap
df['p_gentoo'] = p_gentoo
df
| species | body_mass_g | bill_depth_mm | p_m_given_c | p_m_given_g | p_chinstrap | p_gentoo | |
|---|---|---|---|---|---|---|---|
| 152 | Gentoo | 4500.0 | 13.2 | 1.386413e-04 | 0.000395 | 0.363636 | 0.636364 |
| 153 | Gentoo | 5700.0 | 16.3 | 1.767096e-09 | 0.000381 | 0.363636 | 0.636364 |
| 154 | Gentoo | 4450.0 | 14.1 | 1.788899e-04 | 0.000349 | 0.363636 | 0.636364 |
| 155 | Gentoo | 5700.0 | 15.2 | 1.767096e-09 | 0.000381 | 0.363636 | 0.636364 |
| 156 | Gentoo | 5400.0 | 14.5 | 7.477526e-08 | 0.000661 | 0.363636 | 0.636364 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 339 | Chinstrap | 4000.0 | 19.8 | 8.186991e-04 | 0.000073 | 0.363636 | 0.636364 |
| 340 | Chinstrap | 3400.0 | 18.1 | 7.143042e-04 | 0.000003 | 0.363636 | 0.636364 |
| 341 | Chinstrap | 3775.0 | 18.2 | 1.039432e-03 | 0.000025 | 0.363636 | 0.636364 |
| 342 | Chinstrap | 4100.0 | 19.0 | 6.585033e-04 | 0.000111 | 0.363636 | 0.636364 |
| 343 | Chinstrap | 3775.0 | 18.7 | 1.039432e-03 | 0.000025 | 0.363636 | 0.636364 |
187 rows × 7 columns
In our model, each individual mass can only come about in one of two situations: the penguin is a Chinstrap, or the penguin is a Gentoo. That is, the mass can only come about in these situations:
or:
But:
and
Let’s calculate those now:
df['p_m_and_c'] = df['p_m_given_c'] * df['p_chinstrap']
df['p_m_and_g'] = df['p_m_given_g'] * df['p_gentoo']
\(P(m \text{ and } C)\), \(P(m \text{ and } G)\) are mutually exclusive, so we add the probabilities to give the probability of getting a particular mass value \(m\):
df['p_mass'] = df['p_m_and_c'] + df['p_m_and_g']
df
| species | body_mass_g | bill_depth_mm | p_m_given_c | p_m_given_g | p_chinstrap | p_gentoo | p_m_and_c | p_m_and_g | p_mass | |
|---|---|---|---|---|---|---|---|---|---|---|
| 152 | Gentoo | 4500.0 | 13.2 | 1.386413e-04 | 0.000395 | 0.363636 | 0.636364 | 5.041503e-05 | 0.000252 | 0.000302 |
| 153 | Gentoo | 5700.0 | 16.3 | 1.767096e-09 | 0.000381 | 0.363636 | 0.636364 | 6.425805e-10 | 0.000243 | 0.000243 |
| 154 | Gentoo | 4450.0 | 14.1 | 1.788899e-04 | 0.000349 | 0.363636 | 0.636364 | 6.505088e-05 | 0.000222 | 0.000287 |
| 155 | Gentoo | 5700.0 | 15.2 | 1.767096e-09 | 0.000381 | 0.363636 | 0.636364 | 6.425805e-10 | 0.000243 | 0.000243 |
| 156 | Gentoo | 5400.0 | 14.5 | 7.477526e-08 | 0.000661 | 0.363636 | 0.636364 | 2.719100e-08 | 0.000421 | 0.000421 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 339 | Chinstrap | 4000.0 | 19.8 | 8.186991e-04 | 0.000073 | 0.363636 | 0.636364 | 2.977088e-04 | 0.000046 | 0.000344 |
| 340 | Chinstrap | 3400.0 | 18.1 | 7.143042e-04 | 0.000003 | 0.363636 | 0.636364 | 2.597470e-04 | 0.000002 | 0.000261 |
| 341 | Chinstrap | 3775.0 | 18.2 | 1.039432e-03 | 0.000025 | 0.363636 | 0.636364 | 3.779754e-04 | 0.000016 | 0.000394 |
| 342 | Chinstrap | 4100.0 | 19.0 | 6.585033e-04 | 0.000111 | 0.363636 | 0.636364 | 2.394557e-04 | 0.000071 | 0.000310 |
| 343 | Chinstrap | 3775.0 | 18.7 | 1.039432e-03 | 0.000025 | 0.363636 | 0.636364 | 3.779754e-04 | 0.000016 | 0.000394 |
187 rows × 10 columns
How we have everything we need to work out the posterior probability that a penguin is a Chinstrap, given the weight of the penguin:
df['p_c_given_m'] = (df['p_m_given_c'] * df['p_chinstrap']) / df['p_mass']
df
| species | body_mass_g | bill_depth_mm | p_m_given_c | p_m_given_g | p_chinstrap | p_gentoo | p_m_and_c | p_m_and_g | p_mass | p_c_given_m | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 152 | Gentoo | 4500.0 | 13.2 | 1.386413e-04 | 0.000395 | 0.363636 | 0.636364 | 5.041503e-05 | 0.000252 | 0.000302 | 0.166976 |
| 153 | Gentoo | 5700.0 | 16.3 | 1.767096e-09 | 0.000381 | 0.363636 | 0.636364 | 6.425805e-10 | 0.000243 | 0.000243 | 0.000003 |
| 154 | Gentoo | 4450.0 | 14.1 | 1.788899e-04 | 0.000349 | 0.363636 | 0.636364 | 6.505088e-05 | 0.000222 | 0.000287 | 0.226439 |
| 155 | Gentoo | 5700.0 | 15.2 | 1.767096e-09 | 0.000381 | 0.363636 | 0.636364 | 6.425805e-10 | 0.000243 | 0.000243 | 0.000003 |
| 156 | Gentoo | 5400.0 | 14.5 | 7.477526e-08 | 0.000661 | 0.363636 | 0.636364 | 2.719100e-08 | 0.000421 | 0.000421 | 0.000065 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 339 | Chinstrap | 4000.0 | 19.8 | 8.186991e-04 | 0.000073 | 0.363636 | 0.636364 | 2.977088e-04 | 0.000046 | 0.000344 | 0.865038 |
| 340 | Chinstrap | 3400.0 | 18.1 | 7.143042e-04 | 0.000003 | 0.363636 | 0.636364 | 2.597470e-04 | 0.000002 | 0.000261 | 0.993768 |
| 341 | Chinstrap | 3775.0 | 18.2 | 1.039432e-03 | 0.000025 | 0.363636 | 0.636364 | 3.779754e-04 | 0.000016 | 0.000394 | 0.960216 |
| 342 | Chinstrap | 4100.0 | 19.0 | 6.585033e-04 | 0.000111 | 0.363636 | 0.636364 | 2.394557e-04 | 0.000071 | 0.000310 | 0.772415 |
| 343 | Chinstrap | 3775.0 | 18.7 | 1.039432e-03 | 0.000025 | 0.363636 | 0.636364 | 3.779754e-04 | 0.000016 | 0.000394 | 0.960216 |
187 rows × 11 columns
Likewise:
df['p_g_given_m'] = (df['p_m_given_g'] * df['p_gentoo']) / df['p_mass']
df
| species | body_mass_g | bill_depth_mm | p_m_given_c | p_m_given_g | p_chinstrap | p_gentoo | p_m_and_c | p_m_and_g | p_mass | p_c_given_m | p_g_given_m | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 152 | Gentoo | 4500.0 | 13.2 | 1.386413e-04 | 0.000395 | 0.363636 | 0.636364 | 5.041503e-05 | 0.000252 | 0.000302 | 0.166976 | 0.833024 |
| 153 | Gentoo | 5700.0 | 16.3 | 1.767096e-09 | 0.000381 | 0.363636 | 0.636364 | 6.425805e-10 | 0.000243 | 0.000243 | 0.000003 | 0.999997 |
| 154 | Gentoo | 4450.0 | 14.1 | 1.788899e-04 | 0.000349 | 0.363636 | 0.636364 | 6.505088e-05 | 0.000222 | 0.000287 | 0.226439 | 0.773561 |
| 155 | Gentoo | 5700.0 | 15.2 | 1.767096e-09 | 0.000381 | 0.363636 | 0.636364 | 6.425805e-10 | 0.000243 | 0.000243 | 0.000003 | 0.999997 |
| 156 | Gentoo | 5400.0 | 14.5 | 7.477526e-08 | 0.000661 | 0.363636 | 0.636364 | 2.719100e-08 | 0.000421 | 0.000421 | 0.000065 | 0.999935 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 339 | Chinstrap | 4000.0 | 19.8 | 8.186991e-04 | 0.000073 | 0.363636 | 0.636364 | 2.977088e-04 | 0.000046 | 0.000344 | 0.865038 | 0.134962 |
| 340 | Chinstrap | 3400.0 | 18.1 | 7.143042e-04 | 0.000003 | 0.363636 | 0.636364 | 2.597470e-04 | 0.000002 | 0.000261 | 0.993768 | 0.006232 |
| 341 | Chinstrap | 3775.0 | 18.2 | 1.039432e-03 | 0.000025 | 0.363636 | 0.636364 | 3.779754e-04 | 0.000016 | 0.000394 | 0.960216 | 0.039784 |
| 342 | Chinstrap | 4100.0 | 19.0 | 6.585033e-04 | 0.000111 | 0.363636 | 0.636364 | 2.394557e-04 | 0.000071 | 0.000310 | 0.772415 | 0.227585 |
| 343 | Chinstrap | 3775.0 | 18.7 | 1.039432e-03 | 0.000025 | 0.363636 | 0.636364 | 3.779754e-04 | 0.000016 | 0.000394 | 0.960216 | 0.039784 |
187 rows × 12 columns
df['bayes_predictions'] = np.where(
df['p_c_given_m'] > df['p_g_given_m'],
'Chinstrap', 'Gentoo')
df
| species | body_mass_g | bill_depth_mm | p_m_given_c | p_m_given_g | p_chinstrap | p_gentoo | p_m_and_c | p_m_and_g | p_mass | p_c_given_m | p_g_given_m | bayes_predictions | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 152 | Gentoo | 4500.0 | 13.2 | 1.386413e-04 | 0.000395 | 0.363636 | 0.636364 | 5.041503e-05 | 0.000252 | 0.000302 | 0.166976 | 0.833024 | Gentoo |
| 153 | Gentoo | 5700.0 | 16.3 | 1.767096e-09 | 0.000381 | 0.363636 | 0.636364 | 6.425805e-10 | 0.000243 | 0.000243 | 0.000003 | 0.999997 | Gentoo |
| 154 | Gentoo | 4450.0 | 14.1 | 1.788899e-04 | 0.000349 | 0.363636 | 0.636364 | 6.505088e-05 | 0.000222 | 0.000287 | 0.226439 | 0.773561 | Gentoo |
| 155 | Gentoo | 5700.0 | 15.2 | 1.767096e-09 | 0.000381 | 0.363636 | 0.636364 | 6.425805e-10 | 0.000243 | 0.000243 | 0.000003 | 0.999997 | Gentoo |
| 156 | Gentoo | 5400.0 | 14.5 | 7.477526e-08 | 0.000661 | 0.363636 | 0.636364 | 2.719100e-08 | 0.000421 | 0.000421 | 0.000065 | 0.999935 | Gentoo |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 339 | Chinstrap | 4000.0 | 19.8 | 8.186991e-04 | 0.000073 | 0.363636 | 0.636364 | 2.977088e-04 | 0.000046 | 0.000344 | 0.865038 | 0.134962 | Chinstrap |
| 340 | Chinstrap | 3400.0 | 18.1 | 7.143042e-04 | 0.000003 | 0.363636 | 0.636364 | 2.597470e-04 | 0.000002 | 0.000261 | 0.993768 | 0.006232 | Chinstrap |
| 341 | Chinstrap | 3775.0 | 18.2 | 1.039432e-03 | 0.000025 | 0.363636 | 0.636364 | 3.779754e-04 | 0.000016 | 0.000394 | 0.960216 | 0.039784 | Chinstrap |
| 342 | Chinstrap | 4100.0 | 19.0 | 6.585033e-04 | 0.000111 | 0.363636 | 0.636364 | 2.394557e-04 | 0.000071 | 0.000310 | 0.772415 | 0.227585 | Chinstrap |
| 343 | Chinstrap | 3775.0 | 18.7 | 1.039432e-03 | 0.000025 | 0.363636 | 0.636364 | 3.779754e-04 | 0.000016 | 0.000394 | 0.960216 | 0.039784 | Chinstrap |
187 rows × 13 columns
But - wait - for our Bayes predictions, we don’t actually need the \(P(\text{mass})\) value. Why? Because we can calculate the ratio of the posterior probabilities like this:
likelihood_ratio = ((df['p_m_given_c'] * df['p_chinstrap']) /
(df['p_m_given_g'] * df['p_gentoo']))
# From the divide-out trick above, this gives us the same as:
np.allclose(likelihood_ratio, df['p_c_given_m'] / df['p_g_given_m'])
True
Notice we have divided out the \(P(\text{mass})\) in this ratio. Notice too that ratios above 1 mean more likely Chinstrap, and less than one mean less likely Chinstrap, more likely Gentoo. So we get the same predictions from the likelihood as we would from the full posterior probabilities:
likelihood_predictions = np.where(
likelihood_ratio > 1,
'Chinstrap', 'Gentoo')
np.all(df['bayes_predictions'] == likelihood_predictions)
np.True_
As you may remember from the logit transform in logistic regression, we can also get probabilities from the likelihood ratios, with:
# Probability from likelihood ratio
p_from_lr = likelihood_ratio / (1 + likelihood_ratio)
p_from_lr
152 0.166976
153 0.000003
154 0.226439
155 0.000003
156 0.000065
...
339 0.865038
340 0.993768
341 0.960216
342 0.772415
343 0.960216
Length: 187, dtype: float64
Here is the same process, using the Scikit-learn library.
from sklearn.naive_bayes import GaussianNB
model = GaussianNB()
model.fit(df[['body_mass_g']], df['species']);
model.class_prior_
array([0.36363636, 0.63636364])
df['sklearn_predictions'] = model.predict(df[['body_mass_g']])
df
| species | body_mass_g | bill_depth_mm | p_m_given_c | p_m_given_g | p_chinstrap | p_gentoo | p_m_and_c | p_m_and_g | p_mass | p_c_given_m | p_g_given_m | bayes_predictions | sklearn_predictions | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 152 | Gentoo | 4500.0 | 13.2 | 1.386413e-04 | 0.000395 | 0.363636 | 0.636364 | 5.041503e-05 | 0.000252 | 0.000302 | 0.166976 | 0.833024 | Gentoo | Gentoo |
| 153 | Gentoo | 5700.0 | 16.3 | 1.767096e-09 | 0.000381 | 0.363636 | 0.636364 | 6.425805e-10 | 0.000243 | 0.000243 | 0.000003 | 0.999997 | Gentoo | Gentoo |
| 154 | Gentoo | 4450.0 | 14.1 | 1.788899e-04 | 0.000349 | 0.363636 | 0.636364 | 6.505088e-05 | 0.000222 | 0.000287 | 0.226439 | 0.773561 | Gentoo | Gentoo |
| 155 | Gentoo | 5700.0 | 15.2 | 1.767096e-09 | 0.000381 | 0.363636 | 0.636364 | 6.425805e-10 | 0.000243 | 0.000243 | 0.000003 | 0.999997 | Gentoo | Gentoo |
| 156 | Gentoo | 5400.0 | 14.5 | 7.477526e-08 | 0.000661 | 0.363636 | 0.636364 | 2.719100e-08 | 0.000421 | 0.000421 | 0.000065 | 0.999935 | Gentoo | Gentoo |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 339 | Chinstrap | 4000.0 | 19.8 | 8.186991e-04 | 0.000073 | 0.363636 | 0.636364 | 2.977088e-04 | 0.000046 | 0.000344 | 0.865038 | 0.134962 | Chinstrap | Chinstrap |
| 340 | Chinstrap | 3400.0 | 18.1 | 7.143042e-04 | 0.000003 | 0.363636 | 0.636364 | 2.597470e-04 | 0.000002 | 0.000261 | 0.993768 | 0.006232 | Chinstrap | Chinstrap |
| 341 | Chinstrap | 3775.0 | 18.2 | 1.039432e-03 | 0.000025 | 0.363636 | 0.636364 | 3.779754e-04 | 0.000016 | 0.000394 | 0.960216 | 0.039784 | Chinstrap | Chinstrap |
| 342 | Chinstrap | 4100.0 | 19.0 | 6.585033e-04 | 0.000111 | 0.363636 | 0.636364 | 2.394557e-04 | 0.000071 | 0.000310 | 0.772415 | 0.227585 | Chinstrap | Chinstrap |
| 343 | Chinstrap | 3775.0 | 18.7 | 1.039432e-03 | 0.000025 | 0.363636 | 0.636364 | 3.779754e-04 | 0.000016 | 0.000394 | 0.960216 | 0.039784 | Chinstrap | Chinstrap |
187 rows × 14 columns
Scikit-learn gives the same predictions as we do:
np.all(df['bayes_predictions'] == df['sklearn_predictions'])
np.True_
Scikit-learn also calculates the posterior probabilities:
# The predict_proba values are just the posteriors.
skl_posteriors = model.predict_proba(df[['body_mass_g']])
# They are the same as ours, with calculation error.
our_posteriors = df[['p_c_given_m', 'p_g_given_m']]
assert np.allclose(skl_posteriors, our_posteriors)
The score is the proportion of labels we predicted correctly:
model.score(df[['body_mass_g']], df['species'])
0.9358288770053476
Here we calculated the same value by hand(ish):
np.mean(df['sklearn_predictions'] == df['species'])
np.float64(0.9358288770053476)
Putting the naïve into “naïve Bayes”#
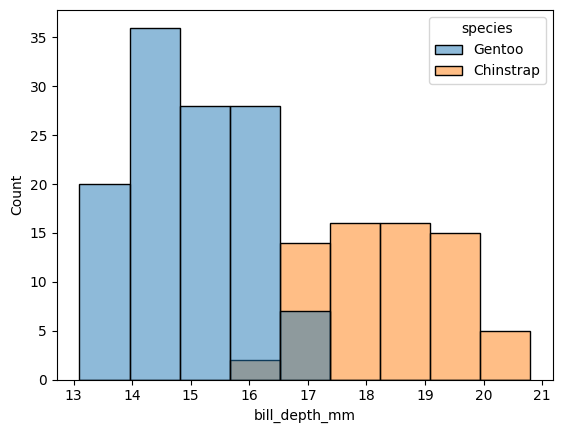
Now, let’s also consider the bill_depth_mm values:
sns.histplot(data=df, x="bill_depth_mm", hue="species")
<Axes: xlabel='bill_depth_mm', ylabel='Count'>
Make the distributions for bill depth:
bd_by_species = by_species['bill_depth_mm']
bd_params = bd_by_species.agg(mean='mean', std=lambda x : np.std(x))
# The distributions
pd_dists = {}
species = ['Chinstrap', 'Gentoo']
for name in species:
pd_dists[name] = sps.norm(bd_params.loc[name, 'mean'],
bd_params.loc[name, 'std'])
pd_dists
{'Chinstrap': <scipy.stats._distn_infrastructure.rv_continuous_frozen at 0x7f5ed1a22990>,
'Gentoo': <scipy.stats._distn_infrastructure.rv_continuous_frozen at 0x7f5ed8674f90>}
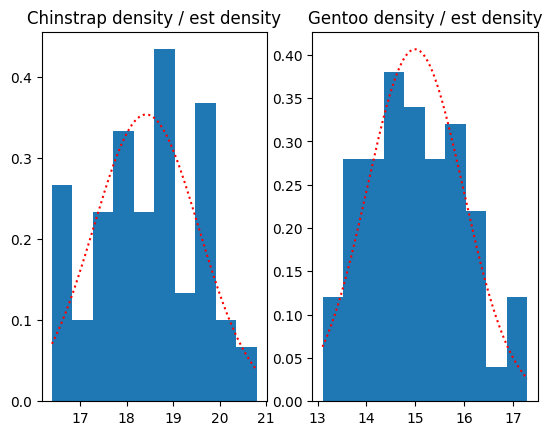
Plot the actual and estimated density, using the Gaussian approximations:
fig, axes = plt.subplots(1, 2)
for i, name in enumerate(species):
axis = axes[i]
bds = bd_by_species.get_group(name)
x = np.linspace(bds.min(), bds.max(), 1000)
axis.hist(bds, density=True)
axis.plot(x, pd_dists[name].pdf(x), 'r:');
axis.set_title(f'{name} density / est density')
We can also think of these distributions in two dimensions, along with the mass:
sns.scatterplot(df, x='body_mass_g', y='bill_depth_mm',
hue='species')
<Axes: xlabel='body_mass_g', ylabel='bill_depth_mm'>
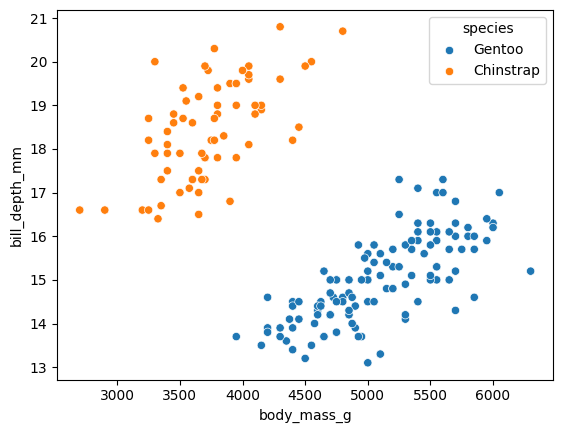
Question - is the bill depth independent (in the sense of probability) from the body mass? That is - if I know the body mass, do I know anything more about the bill depth?
For the moment - let’s say “yes” (it’s independent). That’s naïve. And that’s where “naïve” Bayes reaches the name of the technique.
Using independence, we can calculate the probability of a combination of a particular body mass and bill depth value.
Call \(P(d)\) the probability of a particular bill depth value. Naïve Bayes estimates the probability of the combination with:
Let’s calculate the new probabilities we need:
df['p_bd_given_c'] = pd_dists['Chinstrap'].pdf(df['bill_depth_mm'])
df['p_bd_given_g'] = pd_dists['Gentoo'].pdf(df['bill_depth_mm'])
df
| species | body_mass_g | bill_depth_mm | p_m_given_c | p_m_given_g | p_chinstrap | p_gentoo | p_m_and_c | p_m_and_g | p_mass | p_c_given_m | p_g_given_m | bayes_predictions | sklearn_predictions | p_bd_given_c | p_bd_given_g | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 152 | Gentoo | 4500.0 | 13.2 | 1.386413e-04 | 0.000395 | 0.363636 | 0.636364 | 5.041503e-05 | 0.000252 | 0.000302 | 0.166976 | 0.833024 | Gentoo | Gentoo | 0.000008 | 0.076169 |
| 153 | Gentoo | 5700.0 | 16.3 | 1.767096e-09 | 0.000381 | 0.363636 | 0.636364 | 6.425805e-10 | 0.000243 | 0.000243 | 0.000003 | 0.999997 | Gentoo | Gentoo | 0.060282 | 0.168353 |
| 154 | Gentoo | 4450.0 | 14.1 | 1.788899e-04 | 0.000349 | 0.363636 | 0.636364 | 6.505088e-05 | 0.000222 | 0.000287 | 0.226439 | 0.773561 | Gentoo | Gentoo | 0.000228 | 0.267777 |
| 155 | Gentoo | 5700.0 | 15.2 | 1.767096e-09 | 0.000381 | 0.363636 | 0.636364 | 6.425805e-10 | 0.000243 | 0.000243 | 0.000003 | 0.999997 | Gentoo | Gentoo | 0.005967 | 0.397696 |
| 156 | Gentoo | 5400.0 | 14.5 | 7.477526e-08 | 0.000661 | 0.363636 | 0.636364 | 2.719100e-08 | 0.000421 | 0.000421 | 0.000065 | 0.999935 | Gentoo | Gentoo | 0.000834 | 0.357527 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 339 | Chinstrap | 4000.0 | 19.8 | 8.186991e-04 | 0.000073 | 0.363636 | 0.636364 | 2.977088e-04 | 0.000046 | 0.000344 | 0.865038 | 0.134962 | Chinstrap | Chinstrap | 0.167371 | 0.000003 |
| 340 | Chinstrap | 3400.0 | 18.1 | 7.143042e-04 | 0.000003 | 0.363636 | 0.636364 | 2.597470e-04 | 0.000002 | 0.000261 | 0.993768 | 0.006232 | Chinstrap | Chinstrap | 0.339946 | 0.002751 |
| 341 | Chinstrap | 3775.0 | 18.2 | 1.039432e-03 | 0.000025 | 0.363636 | 0.636364 | 3.779754e-04 | 0.000016 | 0.000394 | 0.960216 | 0.039784 | Chinstrap | Chinstrap | 0.347265 | 0.001984 |
| 342 | Chinstrap | 4100.0 | 19.0 | 6.585033e-04 | 0.000111 | 0.363636 | 0.636364 | 2.394557e-04 | 0.000071 | 0.000310 | 0.772415 | 0.227585 | Chinstrap | Chinstrap | 0.310160 | 0.000100 |
| 343 | Chinstrap | 3775.0 | 18.7 | 1.039432e-03 | 0.000025 | 0.363636 | 0.636364 | 3.779754e-04 | 0.000016 | 0.000394 | 0.960216 | 0.039784 | Chinstrap | Chinstrap | 0.343268 | 0.000331 |
187 rows × 16 columns
The full formula for the posterior using the two measures is:
The naïve version of the formula is:
But - using the likelihood trick above, we no longer have to calculate the denominator, we can just divide it out, to give the following ratio:
both_ratio = ((df['p_m_given_c'] * df['p_bd_given_c'] * df['p_chinstrap']) /
(df['p_m_given_g'] * df['p_bd_given_g'] * df['p_gentoo']))
df['both_predictions'] = np.where(both_ratio > 1, 'Chinstrap', 'Gentoo')
df
| species | body_mass_g | bill_depth_mm | p_m_given_c | p_m_given_g | p_chinstrap | p_gentoo | p_m_and_c | p_m_and_g | p_mass | p_c_given_m | p_g_given_m | bayes_predictions | sklearn_predictions | p_bd_given_c | p_bd_given_g | both_predictions | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 152 | Gentoo | 4500.0 | 13.2 | 1.386413e-04 | 0.000395 | 0.363636 | 0.636364 | 5.041503e-05 | 0.000252 | 0.000302 | 0.166976 | 0.833024 | Gentoo | Gentoo | 0.000008 | 0.076169 | Gentoo |
| 153 | Gentoo | 5700.0 | 16.3 | 1.767096e-09 | 0.000381 | 0.363636 | 0.636364 | 6.425805e-10 | 0.000243 | 0.000243 | 0.000003 | 0.999997 | Gentoo | Gentoo | 0.060282 | 0.168353 | Gentoo |
| 154 | Gentoo | 4450.0 | 14.1 | 1.788899e-04 | 0.000349 | 0.363636 | 0.636364 | 6.505088e-05 | 0.000222 | 0.000287 | 0.226439 | 0.773561 | Gentoo | Gentoo | 0.000228 | 0.267777 | Gentoo |
| 155 | Gentoo | 5700.0 | 15.2 | 1.767096e-09 | 0.000381 | 0.363636 | 0.636364 | 6.425805e-10 | 0.000243 | 0.000243 | 0.000003 | 0.999997 | Gentoo | Gentoo | 0.005967 | 0.397696 | Gentoo |
| 156 | Gentoo | 5400.0 | 14.5 | 7.477526e-08 | 0.000661 | 0.363636 | 0.636364 | 2.719100e-08 | 0.000421 | 0.000421 | 0.000065 | 0.999935 | Gentoo | Gentoo | 0.000834 | 0.357527 | Gentoo |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 339 | Chinstrap | 4000.0 | 19.8 | 8.186991e-04 | 0.000073 | 0.363636 | 0.636364 | 2.977088e-04 | 0.000046 | 0.000344 | 0.865038 | 0.134962 | Chinstrap | Chinstrap | 0.167371 | 0.000003 | Chinstrap |
| 340 | Chinstrap | 3400.0 | 18.1 | 7.143042e-04 | 0.000003 | 0.363636 | 0.636364 | 2.597470e-04 | 0.000002 | 0.000261 | 0.993768 | 0.006232 | Chinstrap | Chinstrap | 0.339946 | 0.002751 | Chinstrap |
| 341 | Chinstrap | 3775.0 | 18.2 | 1.039432e-03 | 0.000025 | 0.363636 | 0.636364 | 3.779754e-04 | 0.000016 | 0.000394 | 0.960216 | 0.039784 | Chinstrap | Chinstrap | 0.347265 | 0.001984 | Chinstrap |
| 342 | Chinstrap | 4100.0 | 19.0 | 6.585033e-04 | 0.000111 | 0.363636 | 0.636364 | 2.394557e-04 | 0.000071 | 0.000310 | 0.772415 | 0.227585 | Chinstrap | Chinstrap | 0.310160 | 0.000100 | Chinstrap |
| 343 | Chinstrap | 3775.0 | 18.7 | 1.039432e-03 | 0.000025 | 0.363636 | 0.636364 | 3.779754e-04 | 0.000016 | 0.000394 | 0.960216 | 0.039784 | Chinstrap | Chinstrap | 0.343268 | 0.000331 | Chinstrap |
187 rows × 17 columns
Accuracy:
np.mean(df['both_predictions'] == df['species'])
np.float64(1.0)
Scikit learn:
both_model = GaussianNB()
both_model.fit(df[['body_mass_g', 'bill_depth_mm']], df['species'])
GaussianNB()In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
GaussianNB()
both_model.score(df[['body_mass_g', 'bill_depth_mm']], df['species'])
1.0
Let’s try the standard test-train split. We “train” our classifier using a random subset of the data:
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
df[['body_mass_g', 'bill_depth_mm']],
df['species'])
X_train
| body_mass_g | bill_depth_mm | |
|---|---|---|
| 210 | 4450.0 | 14.5 |
| 310 | 3600.0 | 18.6 |
| 241 | 5550.0 | 17.0 |
| 208 | 4300.0 | 13.9 |
| 295 | 4400.0 | 18.2 |
| ... | ... | ... |
| 282 | 3250.0 | 18.2 |
| 249 | 4875.0 | 14.6 |
| 219 | 5800.0 | 16.2 |
| 184 | 5050.0 | 14.5 |
| 167 | 5850.0 | 15.7 |
140 rows × 2 columns
Here we fit the various parameters to classify the training data.
test_model = GaussianNB()
test_model.fit(X_train, y_train)
GaussianNB()In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
GaussianNB()
How does Scikit-learn do in classifying the test data (that it has not seen before):
test_model.score(X_test, y_test)
1.0
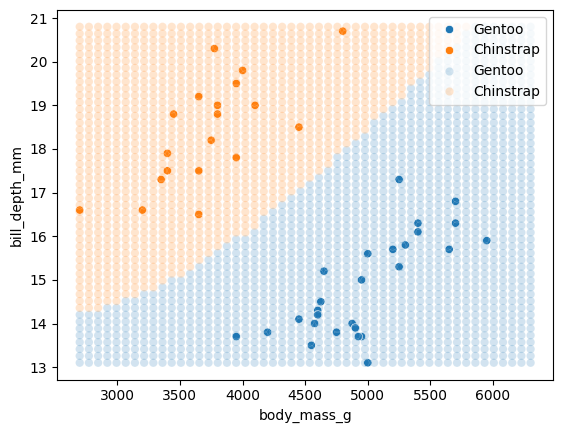
We can look at the decision boundary to see where the model starts seeing Chinstrap and Gentoo penguins, as it moves through the 2D space of parameters (mass and bill depth).
# Make grid of points to classify
all_params = df[['body_mass_g', 'bill_depth_mm']].describe()
bm_x = np.linspace(all_params.loc['min', 'body_mass_g'],
all_params.loc['max', 'body_mass_g'],
50)
bm_y = np.linspace(all_params.loc['min', 'bill_depth_mm'],
all_params.loc['max', 'bill_depth_mm'],
50)
x, y = np.meshgrid(bm_x, bm_y)
xy = np.stack((x.ravel(), y.ravel()), axis=1)
xy_df = pd.DataFrame(xy, columns=X_test.columns)
# Show the classification of the test data.
sns.scatterplot(df.loc[X_test.index],
x='body_mass_g', y='bill_depth_mm',
hue='species')
# Overlay the classification of the grid points.
sns.scatterplot(x=xy[:, 0], y=xy[:, 1],
hue=test_model.predict(xy_df),
palette=sns.color_palette()[:2],
alpha=0.2);